Quick hit demos
These are all miscellaneous features that may be difficult to find in other documentation.
$ operator for model object gets the parameter value
If our model parameters are
Model parameters (N=14):
name value . name value
CL 1 | SEX 0
D1 2 | SEXCL 0.7
F1 1 | SEXVC 0.85
IC50 10 | VC 20
KA 1.2 | WT 70
KIN 100 | WTCL 0.75
KOUT 2 | WTVC 1 we can pick a parameter value with
Or slice off multiple parameters
Model names
For programming with a model object, we can extract the names
$param
[1] "CL" "VC" "KA" "F1" "D1" "WTCL" "WTVC" "SEXCL" "SEXVC"
[10] "KIN" "KOUT" "IC50" "WT" "SEX"
$init
[1] "GUT" "CENT" "RESP"
$capture
[1] "DV" "CP"
$omega
[1] "..."
$sigma
[1] "..."
$omega_labels
$omega_labels[[1]]
[1] "ECL" "EVC" "EKA" "EKOUT"
$sigma_labels
$sigma_labels[[1]]
[1] "EXPO"or get all of the model elements in a list (output not shown)
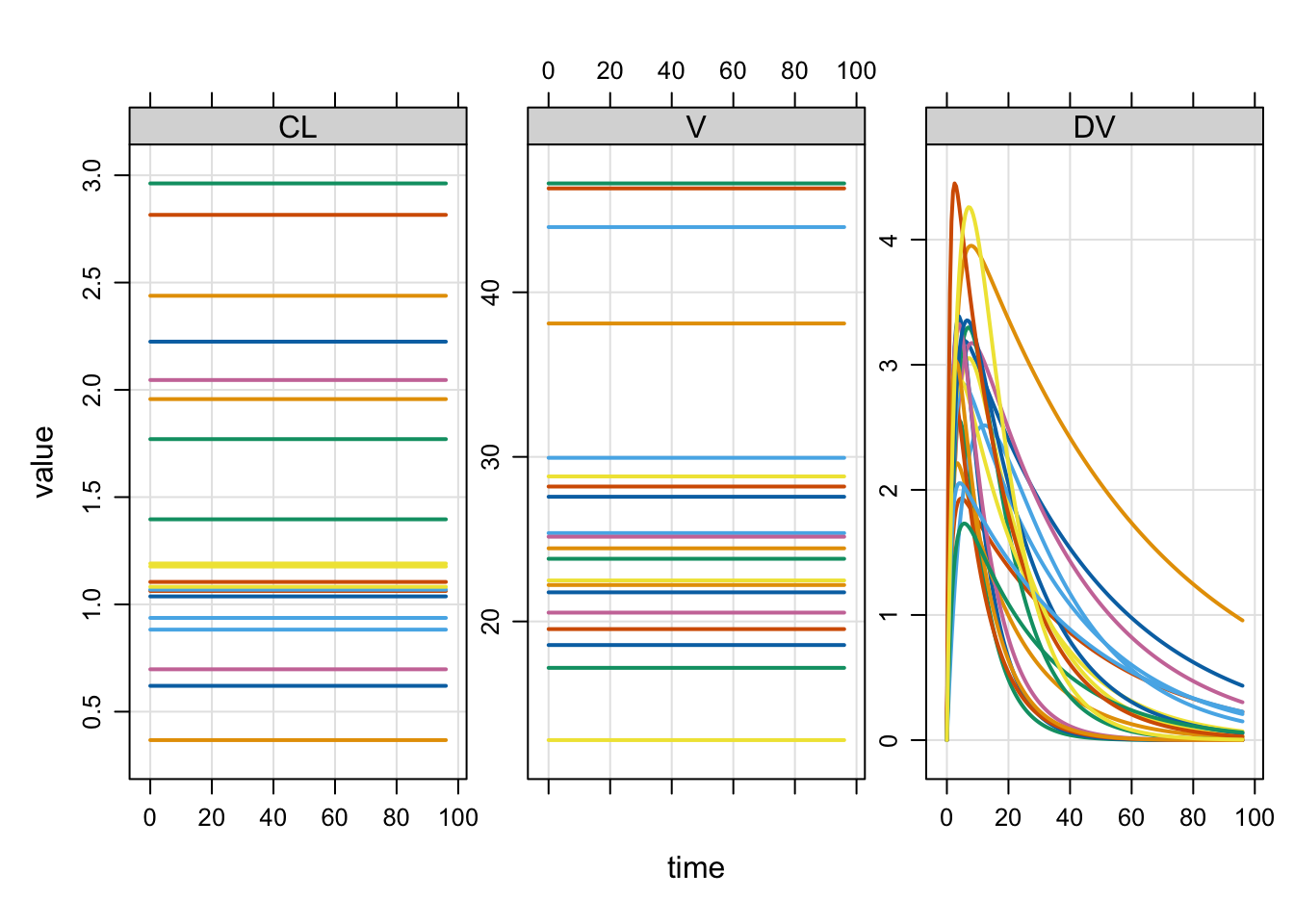
Zero all random effect variances on the fly
If your model has random effects, you can easily and temporarily zero them out.
Building popex ... done.$...
[,1] [,2] [,3]
ECL: 0.3 0.0 0.0
EV: 0.0 0.1 0.0
EKA: 0.0 0.0 0.5It is easy to simulate either with or without the random effects in the simulation: this change can be made on the fly.
Use zero_re to make all random effect variances zero
By default, both OMEGA and SIGMA are zeroed. Check the arguments for zero_re to see how to selectively zero OMEGA or SIGMA.
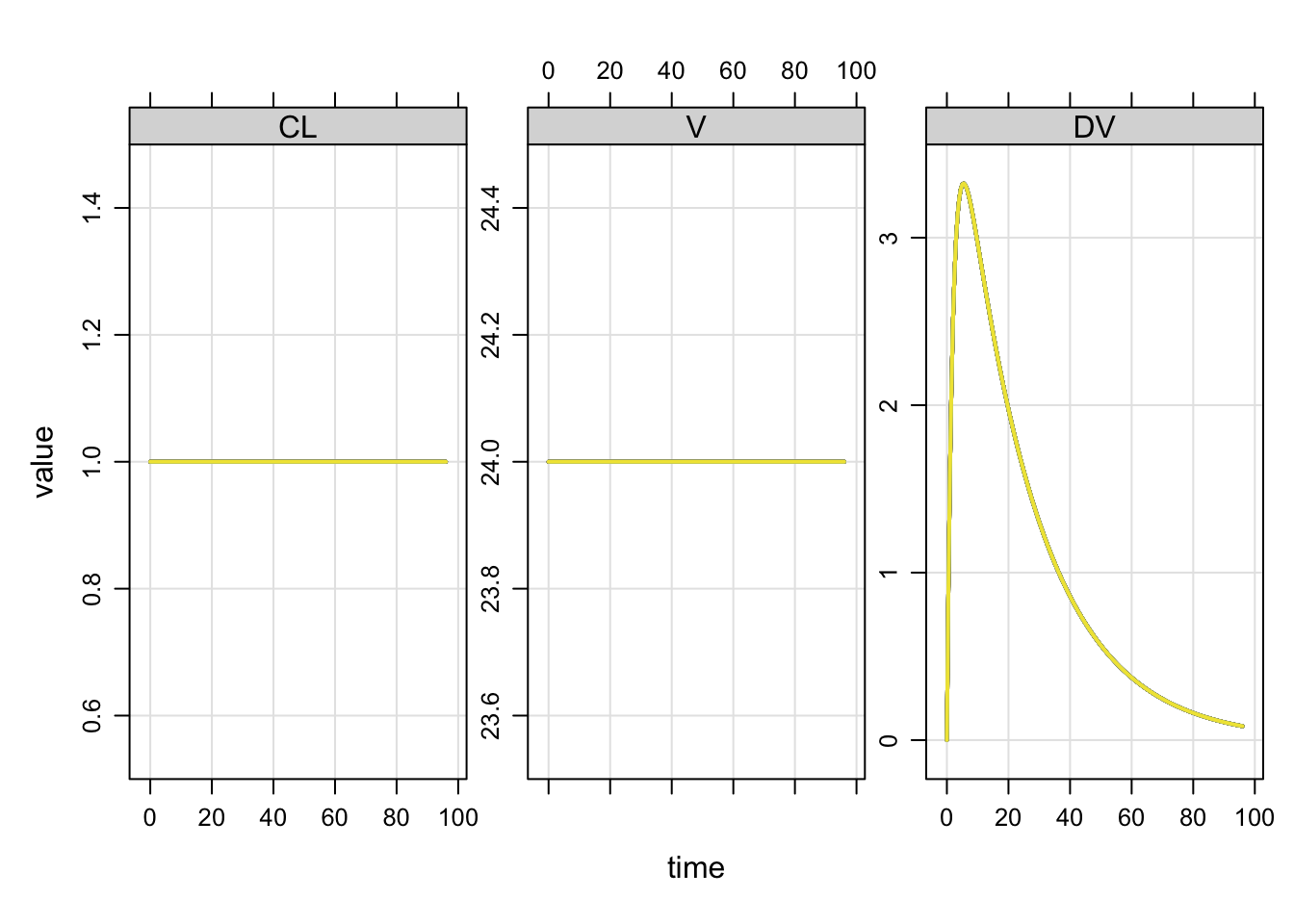
Compare the population output
with
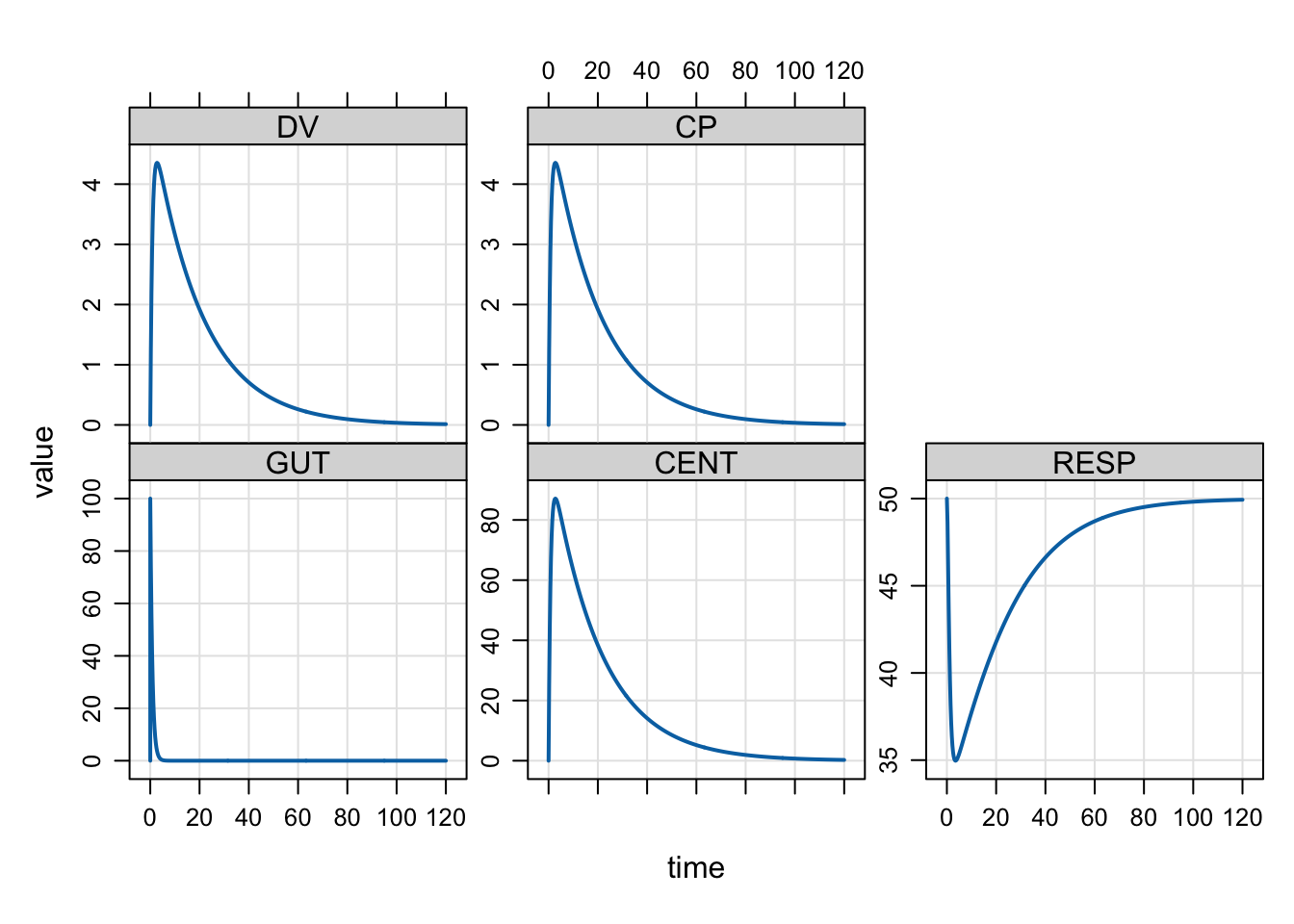
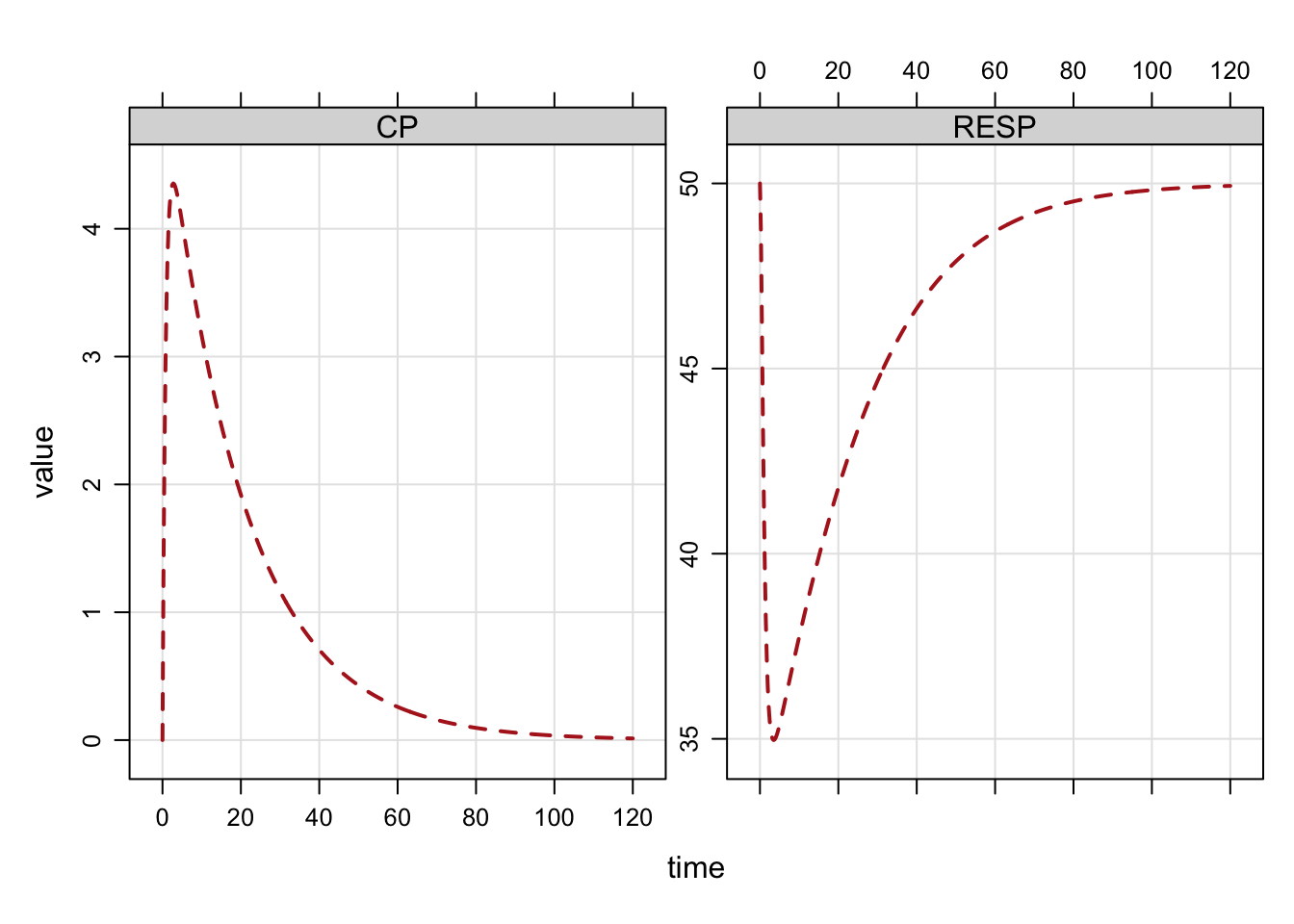
Plot formulae
We commonly plot simulated output with a special plot method. By default, you get all compartments and output variables in the plot.
The plot can be customized with a formula selecting variables to plot. Other arguments
to lattice::xyplot can be passed as well.
Get a data frame of simulated data
By default mrgsolve returns an object of simulated data (and other stuff)
But you can get a data frame with
or